Introduction
What is Bionode?
Modular and universal bioinformatics
Modular | Follows the UNIX philosophy of small tools, each one just trying to do "one thing well". Leverages the Node.JS community and NPM ecosystem for this.
Universal | Can be used anywhere, on a laptop, a server or even a browser! Thanks to being based on JavaScript (JS). Some parts of Bionode also provide a Command Line Interface that allows using it without writing or learning any JS.
Bioinformatics | Aims at leveraging Node.JS Streams to process in a scalable way the huge amount of data being generated by DNA sequencing technologies. However, some parts and lessons learned from this project can be used for any data intensive field (e.g., bionode-watermill).
How did it started?
Bruno Vieira started it in 2014 as a side project of his PhD in Bioinformatics and Population Genomics at WurmLab because he needed to write code for several purposes, and some of it would eventually end up having to be rewritten in JavaScript for some biological web projects. Since he had a background in Node.JS, he decided to just write everything in Node.JS from the start, and also benefit of some of Node.JS' features that seemed very appropriate for orchestrating bioinformatics pipelines. The philosophy of small modules that is present in the Node.JS community also allowed for each piece of code to be quickly published as a module with extras (continuous integration, auto-generated documentation, badges) rather than getting lost as scripts somewhere waiting to be integrated in a bigger monolithic project.
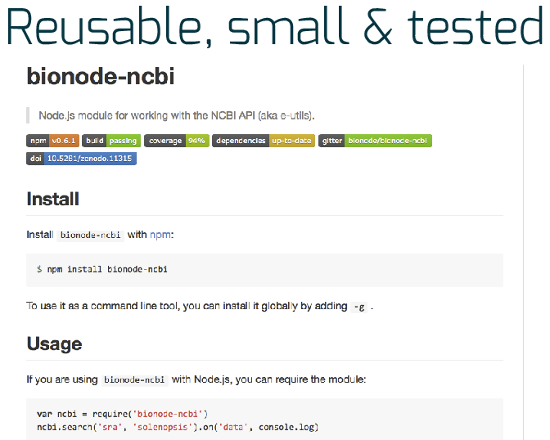 Exemple of a bionode module published on GitHub with quality control badges after the name.
Exemple of a bionode module published on GitHub with quality control badges after the name.
How big is the community?
Bionode has a small community, with a few active developers that contributed new modules.
 Community spread across academic institutions, open source collaborations, and startups.
Community spread across academic institutions, open source collaborations, and startups.
We also organise hackathons and workshops that produce some prototypes of new tools.
 Bionode Hackday at Google Campus London gives rise to five new prototype tools for fetching and processing biological data
Bionode Hackday at Google Campus London gives rise to five new prototype tools for fetching and processing biological data
What kind of tools are you developing?
The overall theme is bioinformatics. Besides that, any tools is welcome. The idea is that if someone is writing JavaScript for biology, and agrees with some of our guidelines, then that code is very welcome in Bionode. Currently there's modules to:
- Retrieve biological data from web resources (e.g., bionode-ncbi)
- Deal with specific bioinformatic data formats (e.g., bionode-fasta)
- Handle sequences (e.g., bionode-seq).
- There's also wrappers around existing bioinformatics tools (e.g., bionode-sam), which obviously only run server side (since those tools are not written in JavaScript) to make it easier to build pipelines in Node.js. However there is some discussion if wrappers are in the scope of Bionode or should we rely on project like the Common Workflow Language (CWL).
- A workflow engine (bionode-watermill) that allows to describe in a clean way complex data processing pipelines with very intricate dependencies and parallelism. It can also be used in fields outside of biology.
- Once we have more basic tools, there might be a move towards genetic algorithms and machine learning.
Current state
You can see a global list of modules on GitHub. Many are still in early development, so check the READMEs and the status badges. Some modules have been requested by the community but not yet developed. On each README you have a link to the documentation of each module, but you can also check a concatenated version of all the documentation at http://doc.bionode.io. You can also have an overview of all the issues at waffle.io. Theres plenty of opportunity for collaboration, and contributors will be coauthors on a paper, so check the "How to contribute" section.
Code produced during events like hackathons is stored under the bionode-hack organisation on GitHub, and are a good way to kickstart a bigger contribution to the project.
Get involved
You can also check our Twitter account for updates or come ask questions in the Gitter chat room. You can also just open an issue on GitHub with your question or idea. Have a look around and come chat to us, there's plenty of ways to get involved, from either the biology side or the computational side.
Collaborations
Dat
Bionode collaborates with Dat, a "Git for data". Dat goals are version control and p2p distribution of big data. The Dat developers have contributed and helped bionode, and both projects follow similar standards. For example, Bionode influenced some of the development decisions in Dat, towards the goal of making Dat the definitive format to share scientific data. Dat influenced Bionode code practices and standards. We also mentored a Google Summer of Code student together.

Open Bioinformatics Foundation
We had in 2016 a Google Summer of Code student, Julian Mazzitelli, with the OBF. He worked full-time for three months and developed a workflow engine with the aim of making it easier to write modular and scalable bioinformatics pipelines. However, his tool is applicable in any data processing workflow in other fields.

BioJS
BioJS is a set of libraries for vizualising biological data on the web. It also provides as registry of biological tools on the web. Bionode and BioJS collaborate to improve compatibility and integration between both projects, for example, by using the same package manager (npm). We also submit Google Summer of Code projects together.
Since both projects are JavaScript and have the word "bio" in the name, people ask frequently what is the difference between them? There are several, but they can be summarized by the fact that Bionode is focusing and "data processing/pipelines" (more backend, even when in a browser) while BioJS is focusing on "visualization". This means that even if the language is the same (which is great for integration) the architecture of each project is very different.
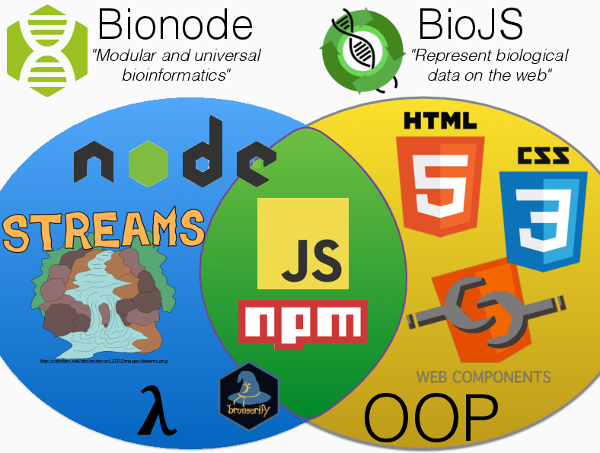 Diagram below with some of the technologies used by each project.
Diagram below with some of the technologies used by each project.
Node.js? Really??
JavaScript is currently the only real "write once, run anywhere" language, and for most cases it's fast enough. Nowadays you can even run very demanding 3D apps compiled to JavaScript in your browser. If you want, you can also run C code in Node.js for speed. The asynchronous and evented nature of Node.js also makes if very interesting for big data pipelines.
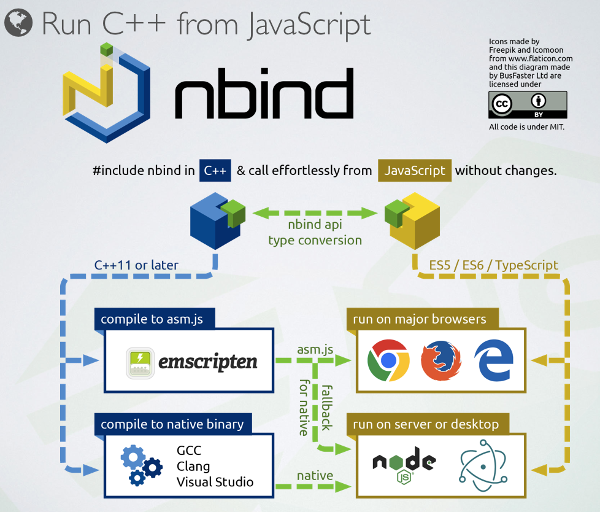 There's a lot of potential for integration between C++ and JavaScript, has shown in the nbind project. We hope in the future to take more advantage of this in Bionode for intensive algorithms.
There's a lot of potential for integration between C++ and JavaScript, has shown in the nbind project. We hope in the future to take more advantage of this in Bionode for intensive algorithms.
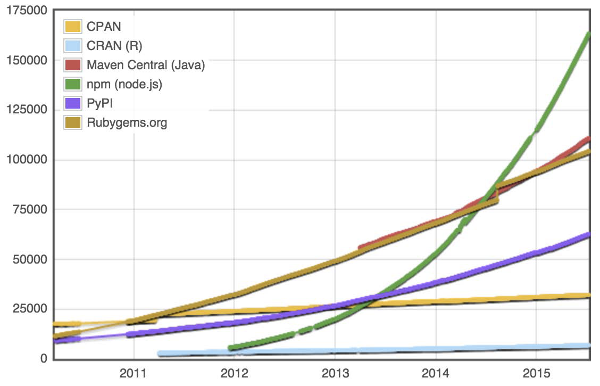 Node.JS is the fastest growing open source community!
Node.JS is the fastest growing open source community!
Sure, it still doesn't have has many bio tools as Python or statistics as R, but that's an opportunity to contribute and get involved in some of the other cool things the Node.js community is making.
Talks and workshops
Here's a list of some of the events we've been: