Pipelines (WIP section)
Update (#MozFest): Try our new bionode-watermill and Julian's beginners tutorial
ncbi
.search('sra', 'Solenopsis invicta')
.pipe(fork1)
.pipe(dat.reads)
fork1
.pipe(tool.extractProperty('expxml.Biosample.id'))
.pipe(ncbi.search('biosample'))
.pipe(dat.samples)
fork1
.pipe(tool.extractProperty('uid'))
.pipe(ncbi.link('sra', 'pubmed'))
.pipe(ncbi.search('pubmed'))
.pipe(fork2)
.pipe(dat.papers)
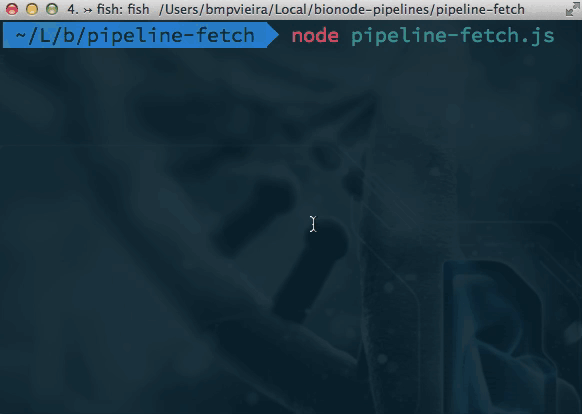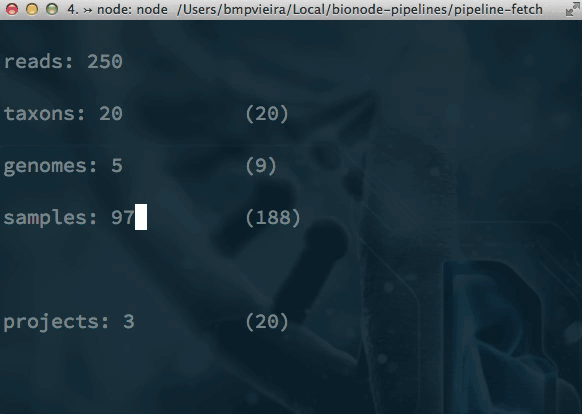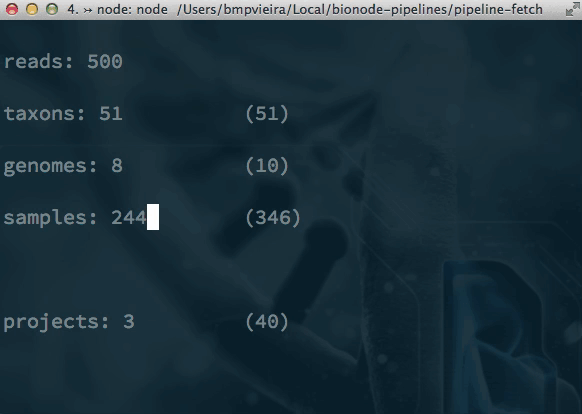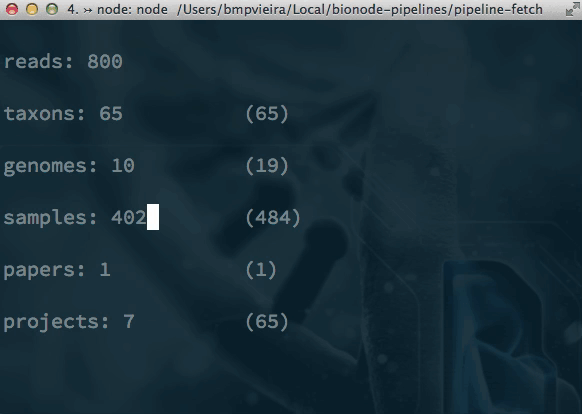
Pipelines and alternatives to Makefiles? bionode-example-dat-gasket get-dat bionode gasket example datproject/datscript groundwater/datscript mafintosh/datscript melaniecebula/datscript ekg/datscript hackfile get-dat workshop
CoffeeScript pipeline
ncbi.search 'genome', 'rodentia'
.pipe ncbi.expand 'assembly'
.pipe ncbi.expand 'tax'
.pipe getLineage()
.pipe ncbi.link 'tax', 'sra'
.pipe ncbi.expand 'sra'
.pipe through.obj (obj, enc, next) ->
async.map obj.sra, expandBiosample, (error, sra)=>
obj.sra = sra
@push obj
next()
pipeline1
ncbi.search genome rodentia
ncbi.expand assembly
ncbi.expand tax
getLineage
ncbi.link tax sra
ncbi.expand sra
stream (obj, next) ->
async.map obj.sra expandBiosample (sra) =>
obj.sra = sra
@push obj
next()